The novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), emerged in late December 2019 from Wuhan, China, and rapidly spread across the world, causing the ongoing coronavirus disease 2019 (COVID-19) pandemic.
Epidemiological studies and sequence-based surveillance have identified multiple SARS-CoV-2 variants with distinct mutations impacting transmissibility, severity of disease, and effectiveness of drugs and vaccines.
The B.1.1.7 variant of SARS-CoV-2 (Alpha), first reported in the United Kingdom, and now classified as a ‘variant of concern,’ is linked to increased transmissibility and disease severity but minimal impact on the monoclonal antibody treatment efficacy.
Several studies have reported an increase in transmissibility in the B.1.1.7 variant with a nearly 90% increase in transmission compared to previously reported variants such as the D614G variant.
However, the increase in mortality related to the B.1.1.7 variant reported earlier in COVID case analysis is questionable. Experimental infections in animal species closely related to humans, such as nonhuman primates, is one way to determine transmissibility and disease severity of emerging variants of SARS-CoV-2.
An intranasal model of SARS-CoV-2 infection to determine differences between the D614G variant and the B.1.1.7 variant of concern
A team of researchers from the US and UK used the AGM intranasal model of SARS-CoV-2 infection to determine differences between a contemporary SARS-CoV-2 D614G variant circulating in summer 2020 and the B.1.1.7 UK variant of concern that emerged in late 2020.
In a paper recently posted to the bioRxiv* preprint server the researchers reported and discussed differences in organ tropism, replication, and shedding between the two viral variants.
The authors used a nasal atomization device to infect African green monkeys with either a contemporary D614G or the UK B.1.1.7 variant to most accurately mimic natural SARS-CoV-2 infection in humans.
Animals belonging to both groups showed minor differences in disease progression following infection with either variant. However, overall signs of the disease were similar, with mild respiratory disease for both the B.1.1.7 and the D614G variants. Although initial studies reported increased disease severity for human B.1.1.7 cases, more recent studies contradicted the earlier claims.
Findings suggest that the B.1.1.7 variant is not associated with increase in disease severity
The study results using a nonhuman primate surrogate model agree with the findings of more recent studies that suggest that the B.1.1.7 variant of concern is not associated with an increase in disease severity.
Although none of the animals in this study developed severe COVID-19, the analysis revealed differences in the two variants in viral replication in the respiratory system. More animals infected with the B.1.1.7 variant had viral RNA and infectious virus in the lower respiratory tract tissues compared to the D614G variant.
This was even more evident during later timepoints suggesting the development of a more potent respiratory component in case of emerging variants of concern.
Moreover, viral RNA shedding in both the nose and oral cavity was also higher in animals infected with the B.1.1.7, which supports human infection data reports showing the B.1.1.7 variant of concern as more transmissible compared to earlier variants.
Distinct tissue or organ tropism of the variants calls for more research given the public health implications
To summarize, the findings of this intranasal AGM COVID-19 surrogate model provide direct empirical data for increased viral replication in the respiratory tissue, with no enhancement of disease severity. Both the variants used caused mild respiratory disease but no significant differences in clinical presentation.
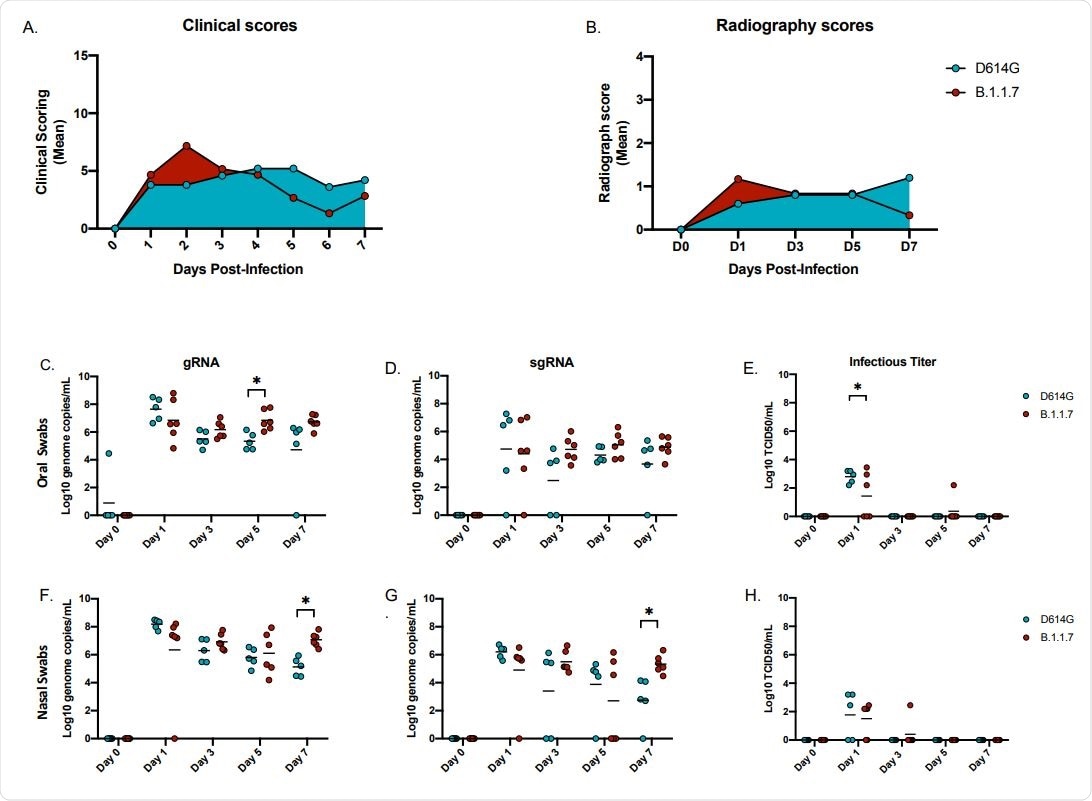
Also, significantly higher levels of viral RNA were detected in upper and lower respiratory tract samples from B.1.1.7 infected animals. Another notable observation of differences among these 2 SARS-CoV-2 variants in terms of distinct viral tissue/organ tropism calls for more research since such tropism could have public health implications concerning viral transmission and disease manifestation.
“In conclusion, our results from the intranasal AGM COVID-19 surrogate model support the most recent data from B.1.1.7 in humans, providing direct empirical data for increased replication in the respiratory tissue, but with no enhancement of disease.”
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- UK B.1.1.7 variant exhibits increased respiratory replication and shedding in nonhuman primates K. Rosenke, F. Feldmann, A. Okumura, F. Hansen, T. Tang-Huau, K. Meade-White, B. Kaza, B.J. Smith, P. W. Hanley, J. Lovaglio, M. A. Jarvis, C. Shaia, H. Feldmann, bioRxiv, 2021.06.11.448134; doi: https://doi.org/10.1101/2021.06.11.448134, https://www.biorxiv.org/content/10.1101/2021.06.11.448134v1
Posted in: Medical Science News | Medical Research News | Disease/Infection News
Tags: Antibody, Coronavirus, Coronavirus Disease COVID-19, Drugs, Efficacy, Genome, Monoclonal Antibody, Mortality, Pandemic, Public Health, Research, Respiratory, Respiratory Disease, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Virus

Written by
Susha Cheriyedath
Susha has a Bachelor of Science (B.Sc.) degree in Chemistry and Master of Science (M.Sc) degree in Biochemistry from the University of Calicut, India. She always had a keen interest in medical and health science. As part of her masters degree, she specialized in Biochemistry, with an emphasis on Microbiology, Physiology, Biotechnology, and Nutrition. In her spare time, she loves to cook up a storm in the kitchen with her super-messy baking experiments.
Source: Read Full Article
