In a recent study conducted at Yale University, USA, scientists have investigated the immunogenicity of mRNA-based coronavirus disease 2019 (COVID-19) vaccine candidates encoding the spike proteins of different severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants. The findings reveal that the vaccines induce robust immune responses against SARS-CoV-2 variants. The study is currently available on the bioRxiv* preprint server.
Background
The continuous emergence of novel SARS-CoV-2 variants with multiple spike mutations has caused a significant reduction in vaccine efficacy worldwide. For instance, the beta variant of SARS-CoV-2, which was first detected in South Africa, has caused a decrease in the effectiveness of the mRNA-based Pfizer/BioNTech COVID-19 vaccine from 90% to 70% by acquiring escape mutations. A similar effect has been observed for the delta variant, which was detected first in India. In addition, there is also grave concern about the recent arrival of the new Omicron variant.
Given the significant negative impacts of viral variants on vaccine efficacy, it is essential to develop next-generation vaccines that can directly target and neutralize more deadly variants of SARS-CoV-2.
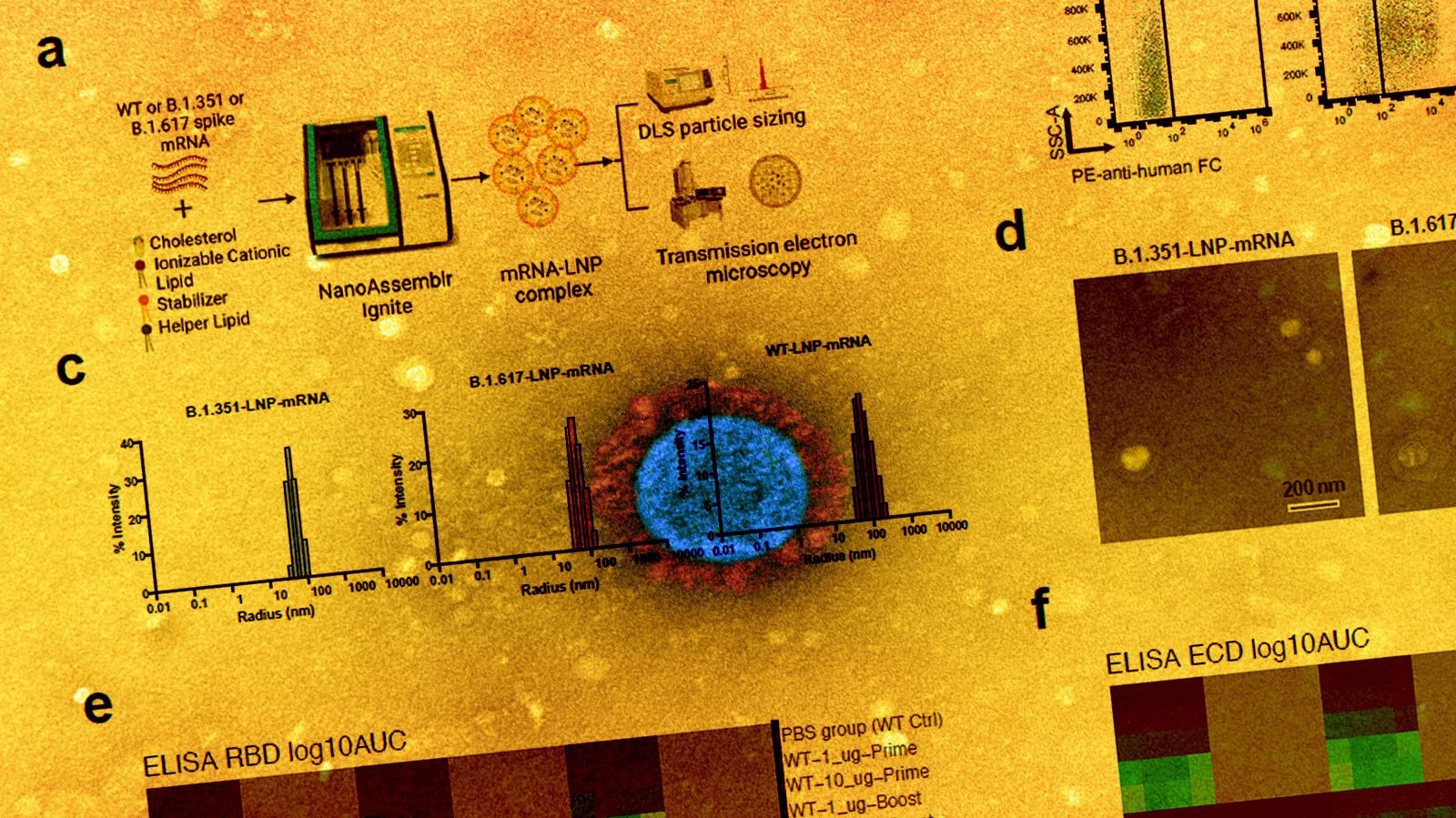
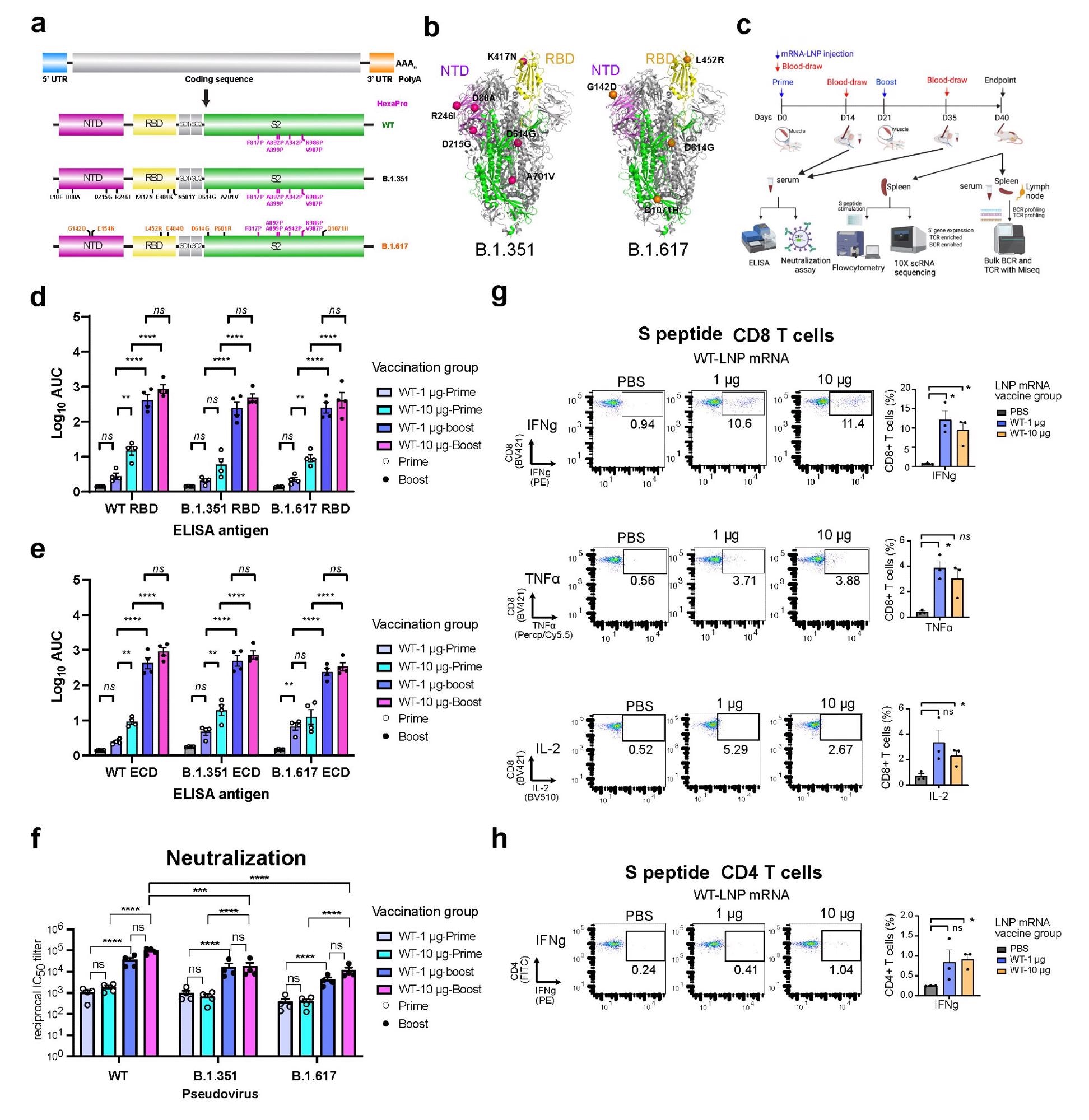
In the current study, the scientists have developed three lipid nanoparticle-formulated mRNA-based vaccine candidates that encode the spike proteins of wild-type SARS-CoV-2 and its variants B.1.351 (Beta) and B.1.617 (14 lineages, including B.1.617.1 “Kappa variant,” B.1.617.2 “Delta variant). In addition, they have specifically investigated the immunogenicity of these vaccine candidates in mice.
They immunized the mice with two doses (prime-boost) of each vaccine intramuscularly at an interval of three weeks. They collected serum samples from the mice two weeks after vaccination to assess antibody response. The mice were sacrificed after 40 days of vaccination and their spleens, lymph nodes, and blood cells were collected for flow cytometry, single-cell transcriptomics, B cell receptor sequencing, and T cell receptor sequencing.
Immune responses to wild-type vaccine
The immunization of mice with wild-type vaccine resulted in strong binding and neutralizing antibody responses against the wild-type virus and all tested variants (B.1.351 and B.1.617). However, the neutralizing potency of vaccinated sera was many folds lower against the variants compared to that against the wild-type virus.
Regarding cellular immunity, the wild-type vaccine showed high efficacy in inducing spike-specific CD8+ T cells secreting interferon-gamma, tumor necrosis factor-alpha, and interleukin-2. Similarly, the vaccine-induced robust spike-specific CD4+ T Cells secreting interferon-gamma.
Immune responses to variant-specific vaccines
Both variant-specific vaccines produced strong antibody responses against wild-type and tested variants, just like the wild-type vaccine. In addition, both variant-specific vaccines induced robust neutralizing antibodies against all tested variants. The binding and neutralizing antibody titers were significantly higher after booster immunization compared to that after prime immunization.
Specifically, the B.1.351-specific vaccine showed similar neutralizing efficacy against all tested viral variants. However, the B.1.617-specific vaccine induced significantly higher antibody titers against the B.1.617 variant compared to that against the wild-type virus and B.1.351 variant.
Regarding cellular immunity, both variant-specific vaccines induced spike-specific CD8+ and CD4+ immune responses similar to the wild-type vaccine.
Immune cell transcriptional landscape following variant-specific vaccination
The findings of single-cell transcriptomics sequencing revealed that both variant-specific vaccines caused a significant increase in activated CD8+ T cells and a slight decrease in natural killer cells. In addition, the B.1.351-specific vaccine caused a slight increase in dendritic cells.
The analysis identified differential expressions of many genes regarding vaccine-induced transcriptomic changes in the B cell and T cell subpopulations. In B cells, differentially expressed genes were associated with B cell activation, immune effector, and immune response. Similarly, in CD4+ and CD8+ T cells, differentially expressed genes were associated with T cell activation, immune effector, and immune response. For both vaccines, the most significantly upregulated genes in B and T cells represented transcription and translation machinery, indicating active lymphocyte activation following vaccination.
B cell and T cell receptor diversity following variant-specific vaccination
The findings of single-cell and bulk B/T cell receptor sequencing revealed a reduction in clonal diversity in the spleen, peripheral blood, and lymph node samples following variant-specific vaccination. These observations indicate that both B.1.351- and B.1.617-specific vaccines cause expansion of a small number of B cell and T cell clones.
Study significance
The study highlights that variant-specific COVID-19 vaccines induce robust humoral and cellular immune responses and are associated with a strong signature of transcriptional and translational machineries in lymphocytes.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Peng L. 2021. Systems immune profiling of variant-specific vaccination against SARS-CoV-2, bioRxiv, https://www.biorxiv.org/content/10.1101/2021.12.02.471028v1
Posted in: Drug Trial News | Medical Research News | Disease/Infection News
Tags: ACE2, Antibodies, Antibody, Assay, B Cell, Blood, CD4, Cell, Coronavirus, Coronavirus Disease COVID-19, Cytokine, Cytometry, Efficacy, Flow Cytometry, Genes, Immune Response, immunity, Immunization, Interferon, Interferon-gamma, Interleukin, Interleukin-2, Intracellular, Lymph Node, Lymph Nodes, Lymphocyte, Nanoparticle, Natural Killer Cells, Necrosis, Protein, Pseudovirus, Receptor, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spleen, Syndrome, Transcription, Transcriptomics, Translation, Tumor, Tumor Necrosis Factor, Vaccine, Virus

Written by
Dr. Sanchari Sinha Dutta
Dr. Sanchari Sinha Dutta is a science communicator who believes in spreading the power of science in every corner of the world. She has a Bachelor of Science (B.Sc.) degree and a Master's of Science (M.Sc.) in biology and human physiology. Following her Master's degree, Sanchari went on to study a Ph.D. in human physiology. She has authored more than 10 original research articles, all of which have been published in world renowned international journals.
Source: Read Full Article
