In a recent study posted to the bioRxiv* pre-print server, researchers at the University of Texas Medical Branch, Pfizer Vaccines Research & Development, and BioNTech assessed the neutralization titers of sera from individuals vaccinated with the BNT162b2 coronavirus disease 2019 (COVID-19) vaccine and previously infected with the Omicron sublineage BA.1.
 Study: Neutralization of Omicron sublineages and Deltacron SARS-CoV-2 by 3 doses of BNT162b2 vaccine or BA.1 infection. Image Credit: peterschreiber media / Shutterstock
Study: Neutralization of Omicron sublineages and Deltacron SARS-CoV-2 by 3 doses of BNT162b2 vaccine or BA.1 infection. Image Credit: peterschreiber media / Shutterstock
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) new variant of concern (VOC) Omicron is evolving into distinct sublineages, BA.1, BA.2, BA.2.12.1, BA.3, BA.4, and BA.5, which are circulating worldwide. In addition, the co-circulation of Omicron and Delta has also led to the emergence of a hybrid variant Deltacron (XD). The newly emerged Omicron sublineages and Deltacron have distinct mutations in their spike (S) glycoproteins. Thus, there is a growing need to examine their susceptibility to the vaccine- or infection-elicited neutralization antibodies (nAbs).
About the study
In the present study, researchers engineered mNeonGreen (mNG) tagged S proteins for SARS-CoV-2 viruses, BA.1-, BA.2-, BA.2.12.1-, BA.3, BA.4/5-, and XD-S. They infected Vero E6 cells with 106 plaque-forming units per milliliter (PFU/ml) of these viruses, similar to the wild-type (wt) USA-WA1/2020 mNG virus.
The team used the recombinant SARS-CoV-2 viruses to determine the 50% fluorescent focus-reduction neutralization titers (FFRNT50) for two panels of human sera. They obtained the first set of serum samples one month after 22 individuals had received dose 3 (PD3) of the BNT162b2 vaccine. The second panel of sera comprised sera samples from 20 individuals who were unvaccinated but contracted an infection from Omicron BA.1 sub-variant. The researchers confirmed the genotype of infecting virus using Sanger sequencing of nasal swabs.
Study findings
The geometric mean titers (GMTs) of the BNT162b2 PD3 immune sera for wt WA1 were the highest at 1335 while the GMTs for BA.1-, BA.2-, BA.2.12.1-, BA.3-, BA.4/5-, and XD-S variants were 393, 298, 315, 216, 103, and 301, respectively. The PD3 sera consistently had neutralization titers below 20 across all SARS-CoV-2 variants.
The S variants of Omicron sublineages, BA.1, BA.2, BA.2.12.1, and XD evaded nAbs-induced by three doses of BNT162b2 at the same propensity. It explains the sequential rise in the prevalence of BA.2.12.1 > BA.2 > BA.1, driven by the differences in SARS-CoV-2 transmissibility or other mechanisms of immune evasion. Also, BNT162b2 PD3 sera much less efficiently neutralized BA.4/5 sublineages. These results implied that the BA.3 sublineage had attenuated fitness, and BA.4 and BA.5 sublineages could soon replace them, leading to a new wave of SARS-CoV-2 infections.
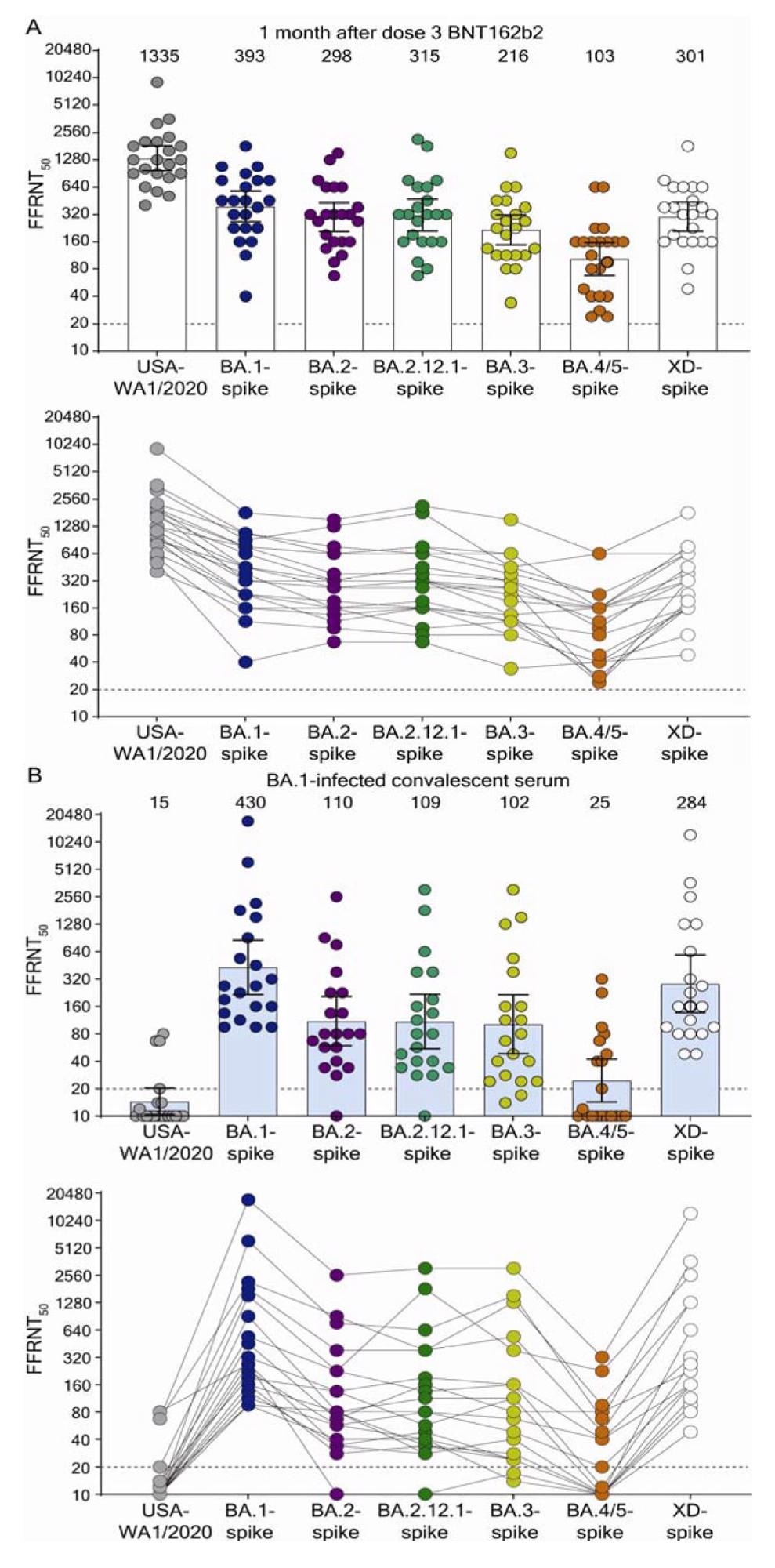
Neutralization by sera collected at 1-month post dose 3 BNT162b2 vaccine (A) and by sera collected from unvaccinated individuals who contracted Omicron BA.1 SARS-CoV-2 (B). Scatterplot of neutralization titers against USA-WA1/2020, Omicron sublineage BA.1-, BA.2-, BA.2.12.1-, BA.3-, BA.4/5-, and Deltacron XD-spike mNG SARS-CoV-2s. Both BNT162b2-vaccinated sera (n=22) and BA.1-infected convalescent sera (n=20) were tested for their FFRNT50s against the variant-spike mNG SARS-CoV-2s. The variant-spike mNG SARS-CoV-2s were produced by engineering the complete variant spike genes into the mNG USA-WA1/2020. Each data point represents the geometric mean FFRNT50 (GMT) obtained with a serum specimen against the indicated virus. Tables S1 and S2 summarize the serum information and FFRNT50s for (A) and (B), respectively. The neutralization titers for individual variant-spike mNG SARS-CoV-2s were determined in two or three independent experiments, each with duplicate assays; the GMTs are presented. The bar heights and the numbers above indicate GMTs. The whiskers indicate 95% confidence intervals. The dotted lines indicate the limit of detection of FFRNT50. Statistical analysis was performed with the use of the Wilcoxon matched-pairs signed-rank test. For the BNT162b2-vaccinated sera in (A), the P values of the GMT differences between USA-WA1/2020 and any variant-spike SARS-CoV-2 are all < 0.0001. For the BA.1-convelescent sera in (B), the P value of GMT difference between BA.1- and XD-spike viruses is 0.0021; the P values of the GMT differences between BA.1- and any other variant-spike viruses (including USA-WA1/2020) are all <0.0001. For both serum panels in (A) and (B), FFRNT50 values with connected lines are presented for individual sera.
The GMTs of neutralizing activity of sera from BA.1-infected individuals was the lowest against wt WA1 at 15. However, GMTs against BA.1-, BA.2-, BA.2.12.1-, BA.3-, BA.4/5-, and XD-S variants were 430, 110, 109, 102, 25, and 284, respectively. However, sera from BA.1-infected individuals could not neutralize all Omicron sublineages and Deltacron XD with similar efficiencies, especially BA.4/5. It is likely that the BA.4/5 S mutation, F486V, helped these sublineages further evade prior infection-induced nAbs.
Limitations and conclusions
An important limitation of the study is that it lacked analyses of T cells and non-neutralizing antibodies that mediate Fraction, crystallizable (Fc)-mediated effector functions. Notably, both types of immunity work in strict coordination to protect against pathogenic infections, such as COVID-19. Nevertheless, the study findings support previous observations on Omicron sublineage neutralization. Therefore, there is an urgent need to update current COVID-19 vaccines, which do not elicit nAbs with the potential to neutralize Omicron BA.4 and BA.5 sublineages.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Neutralization of Omicron sublineages and Deltacron SARS-CoV-2 by 3 doses of BNT162b2 vaccine or BA.1 infection, Chaitanya Kurhade, jing Zou, Hongjie Xia, Mingru Liu, Qi Yang, Mark Cutler, David Cooper, Alexander Muik, Ugur Sahin, Kathrin U. Jansen, Ping Ren, Xuping Xie, Kena A. Swanson, Pei-Yong Shi, bioRxiv pre-print 2022, DOI: https://doi.org/10.1101/2022.06.05.494889, https://www.biorxiv.org/content/10.1101/2022.06.05.494889v2
Posted in: Medical Research News | Disease/Infection News | Pharmaceutical News
Tags: Antibodies, Coronavirus, Coronavirus Disease COVID-19, Efficacy, Genes, immunity, Mutation, Omicron, Research, Respiratory, Sanger sequencing, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Vaccine, Virus

Written by
Neha Mathur
Neha is a digital marketing professional based in Gurugram, India. She has a Master’s degree from the University of Rajasthan with a specialization in Biotechnology in 2008. She has experience in pre-clinical research as part of her research project in The Department of Toxicology at the prestigious Central Drug Research Institute (CDRI), Lucknow, India. She also holds a certification in C++ programming.
Source: Read Full Article
