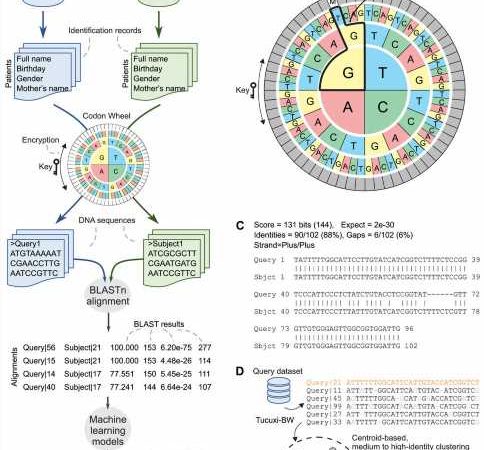
Brazilian researchers have created an innovative and agile computational tool to link and analyze different health databases with millions of patient records. Called Tucuxi-BLAST, the platform encodes identification records in a database, such as patient name, mother’s name and place of birth, using letters that represent the nucleotides in a DNA sequence (A, T, C or G). This “conversion” of individuals to DNA enables accurate record linkage across databases despite typographical errors and other inconsistencies.
The tool can be used in research, epidemiological analysis and public policy formulation.
For example, people who have been vaccinated by the SUS, Brazil’s national health service, can be cross-referenced to other datasets to find vaccinated patients with a specific disease. Even if a vaccination record contains errors or uncompleted fields, Tucuxi-BLAST is able to link it to the same patient in another database because it treats inconsistencies as if they were DNA mutations. Genomics tools routinely need to compare fragments in order to decide whether they are more similar than different and whether to link the base pairs in question. If each individual corresponds to a sequence of letters, data from different repositories can be cross-referenced and linked by the tool.
“The SUS is a valuable source of information for medical and epidemiological research because it stores health data for millions of patients. However, records relating to diseases and other types of data are stored in different databases that don’t always talk to each other. The method we’ve developed is able to effect record linkage accurately and at great speed,” Helder Nakaya, corresponding author of an article on the study published in the journal PeerJ, told Agência FAPESP.
Nakaya is an immunologist affiliated with the University of São Paulo’s School of Pharmaceutical Sciences (FCF-USP), the Albert Einstein Jewish Hospital (HIAE), the Scientific Platform Pasteur-USP, and Todos pela Saúde institute. He also belongs to the Center for Research on Inflammatory Diseases (CRID), one of the Research, Innovation and Dissemination Centers (RIDCs).
Using the tool in practice
Even before the article was published, Tucuxi-BLAST began to be deployed in practice. It was used, for example, to cross-reference four years of data from the Ministry of Health’s Malaria Surveillance System with clinical data from the Dr. Heitor Vieira Dourado Tropical Medicine Foundation (in Manaus, Amazonas state), a branch of Oswaldo Cruz Foundation (Fiocruz), another arm of the ministry.
The result showed that being HIV positive is a risk for Plasmodium vivax malaria patients, representing an additional challenge for public policy. Given the lack of single identifiers, Tucuxi-BLAST used patient name, mother’s name and date of birth. The findings were described in an article published in May 2022 in Scientific Reports.
The study was led by researchers at Amazonas State University (UEA). Nakaya and FCF-USP’s José Deney Alves Araújo, first author of the PeerJ article, also participated. Araújo named the tool Tucuxi in honor of Sotalia fluviatilis, a freshwater dolphin that inhabits the rivers of the Amazon Basin.
BLAST (Basic Local Alignment Search Tool) refers to a suite of programs used in bioinformatics to generate alignments between nucleotides or protein sequences across large databases.
How it works
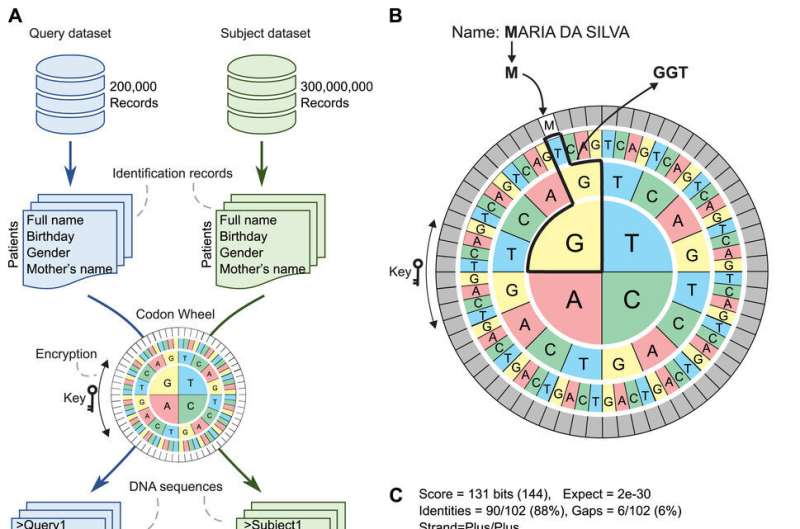
To develop the new method, the scientists translated patient data into DNA sequences using a codon wheel that changed dynamically over different runs without impairing the efficiency of the process. Codons are sequences of three nucleotides that code for a specific amino acid in a DNA or RNA molecule. Codon wheels are used to identify the amino acids encoded by any DNA or RNA codon.
This encoding scheme enabled real-time data encryption, thus providing an additional layer of privacy during the linking process. “It used DNA to encrypt the information and guarantee privacy,” Nakaya said.
The DNA-encoded identification fields were compared using BLAST, and machine learning algorithms automatically classified the final results.
As in comparative genomics, where genes from different genomes are compared to determine common and unique sequences, Tucuxi-BLAST also permits the simultaneous integration of data from multiple administrative databases without the need for complex data pre-processing.
In the study, the group used Tucuxi-BLAST to test and compare a simulated database containing 300 million records, as well as four large administrative databases containing data for real cases of patients infected with different pathogens.
The conclusion was that Tucuxi-BLAST successfully processed record linkages for the largest dataset (200,000 records), despite misspellings and other errors and omissions, in a fifth of the time: 23 hours, compared with 127 hours (five days and seven hours) for the state-of-the-art method.
The researchers set up a website where users can translate words, phrases and names into DNA.
Source: Read Full Article