The nucleus accumbens (NAc) is an area of the brain known to play a key role in regulating a variety of reward-related behaviors, including appetitive and aversive responses, feeding, social interactions and some types of learning. Studies on both humans and animals have also linked NAc dysfunction to several neuropsychiatric disorders, such as depression, anxiety, schizophrenia and substance abuse.
Researchers at Boston Children’s Hospital and Harvard University have recently carried out a study aimed at better understanding the molecular and cellular underpinnings of the NAc. Their paper, published in Nature Neuroscience, outlines one of the most comprehensive molecular and cellular taxonomies of the NAc in the mouse brain constructed to date.
“Consistent with its functional diversity, the NAc establishes complex connections with different up and downstream brain regions,” Renchao Chen, one of the researchers who carried out the study, told Medical Xpress. “However, the cellular basis underlying the anatomic and functional diversity of the NAc is largely unknown.”
The most renowned neuroscientific model of the NAc broadly classifies medium spiny neurons (MSN), the most common neuron type found in this brain region, into two distinct populations. These are the D1 dopamine-receptor-expressing and D2 dopamine-receptor-expressing neuron populations, also referred to as D1 MSN and D2 MSN, respectively.
“Although this conventional model provides a framework for understanding the cellular and circuitry organization of the NAc, it takes little consideration of intra-NAc heterogeneity, and it remains unclear whether distinct neuron subtypes underlie the anatomical and functional diversity of the NAc,” Chen explained. “To answer this question, we sought to generate a molecularly defined and spatially resolved cell taxonomy of the NAc, as it can serve as a framework for integrating its structural and functional diversity.”
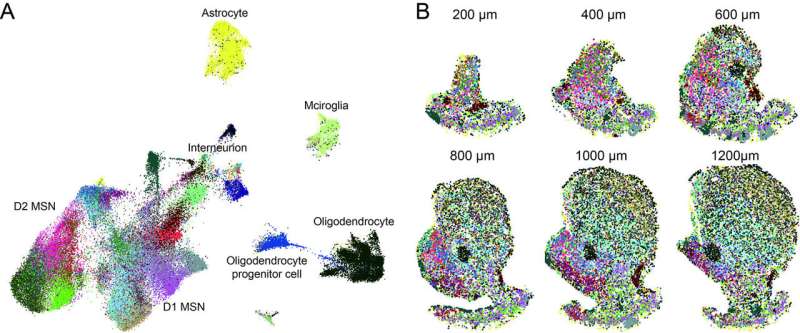
To construct their comprehensive molecular and cellular taxonomy of the NAc, Chen and his colleagues used two complementary techniques often employed by neuroscientists: single-cell RNA sequencing (scRNA-seq) and multiplexed error-robust fluorescence in situ hybridization (MERFISH). ScRNA-seq is a genomic approach that allows researchers to examine the transcriptional (i.e., gene expression) profiles of individual cells. MERFISH, on the other hand, is an imaging technique that can also be used to examine the gene expression profiling of single cells in biological samples.
Using scRNA-seq, the researchers were able to measure the expression of thousands of genes in each of the cells in NAc tissue they extracted from the mouse brain. Subsequently, they divided these cells into different groups or cell types based on their gene expression profiles, to generate a molecular taxonomy of the mouse NAc.
“To further resolve the spatial distribution of these molecularly defined cell types in the brain, we used MERFISH to simultaneously label and count hundreds of different mRNA species with single-molecule resolution in tissue sections, which allowed us to quantify the expression of a panel of 253 genes (chosen based on scRNA-seq) in a serial of thin NAc slices,” Chen explained. “Based on the expression pattern of these genes, we can resolve the identity of each cell and their location in the NAc.”
By integrating the data that they collected using scRNA-seq and MERFISH techniques, Chen and his colleagues were ultimately able to construct a molecularly defined and spatially resolved cell taxonomy of the mouse NAc. Their findings are the first to clearly show that there are many MSN cell subtypes in the NAc, each with distinct gene expression profiles and spatial features.
“We also gathered strong evidence suggesting that gene expression and spatial features of different MSN subtypes underlie the anatomic and functional complexity of the NAc,” Chen said. “The most important implication of our findings is that the different features of NAc, such as neuron subtypes, anatomic sub-regions and functional complexity are correlated with each other and could be integrated into the same framework, which is built on molecularly defined neuron subtypes.”
Overall, the results could have many implications for the study of the NAc and its neural underpinnings. Most notably, they highlight the promise of using molecular tools, such as transgenic mouse lines, to study the cellular and functional diversity of this important brain region.
In the future, this recent study could pave the way for other studies aimed at painting a more exhaustive picture of the mouse and human NAc. Collectively, these works could inform the development of more effective treatment strategies for neuropsychiatric disorders associated with NAc dysfunction.
Source: Read Full Article
