Genetic lineages of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have wreaked havoc by evolving and circulating all over the world. Amongst these, the B.1.617.2 (Delta) variant is the most common lineage found in India and globally. In addition, breakthrough infections with Delta variant have been documented in COVID vaccine recipients and people with prior SARS-CoV-2 infection. It is unclear whether fully vaccinated individuals with two doses of COVID-19 vaccine can transmit the virus.
After two doses of either mRNA or adenovirus-based vaccination, vaccine effectiveness (VE) for the Delta variant is predicted to be around 50% for mild symptomatic infection. Earlier, 155 cases of vaccine-related infections had been documented among frontline Delhi healthcare workers (HCW), most of whom had received one or two doses of the Covishield (ChAdOx1) vaccine, and the majority were immunized >21 days prior to the symptomatic presentation.
According to existing data, vaccinated and unvaccinated individuals have identical viral loads, but the former group elicited a steeper viral decay over time. Also, the likelihood of virus culture was lower in immunized individuals. It's unclear whether those who have a vaccine breakthrough spread it on to other vaccinated people or if the transmission is primarily driven by the unvaccinated individuals.
The Study
A new study published on the medRxiv* preprint server employed in-silico techniques, combining genomic and epidemiological data, to determine possible Delta-variant transmission events between vaccinated healthcare workers (HCWs) in Indian hospitals.
The study design comprised of frontline healthcare workers exhibiting symptoms of SARS-CoV-2, who were tested for viral presence using reverse transcriptase-polymerase chain reaction (RT-PCR).
The bioinformatics and phylogenetic analysis were carried out by obtaining consensus sequences in fasta format from two separate Hospitals in Delhi, India.
Findings
The results found 81 breakthrough infections among 1,100 HCWs in hospital A and 32 breakthrough infections among 4,000 HCWs in hospital B.
B.1.617 was found in 90.7% of the breakthrough infections in doubly vaccinated HCWs. B.1.1.7-like accounted for 5.3% of the total, whereas B.1.538 accounted for 1.3%.
Fever (82.3 %of all cases), cough (43.4 %), myalgia (20.4%), and loss of smell/taste (14.2 %) were the frequently reported symptoms. In hospital A, 66 sequences produced high-quality whole-genome coverage of more than 90%, including 43 cases of breakthrough infection. While in hospital B, 52 samples, including all 32 symptomatic breakthrough infections, were used to obtain high-quality genome sequences.
Numerous introductions into hospitals A and B occurred, with the subsequent intra-hospital transmission. As a result, significant partitioning of the inferred phylogeny was discovered, dividing the sequences into distinct clades.
Mutations to the Delta consensus that were discovered in these cases were detected everywhere over the SARS-CoV-2 genome. Neither of the single-nucleotide polymorphism (SNPs) was found in homoplasic areas of the genome.
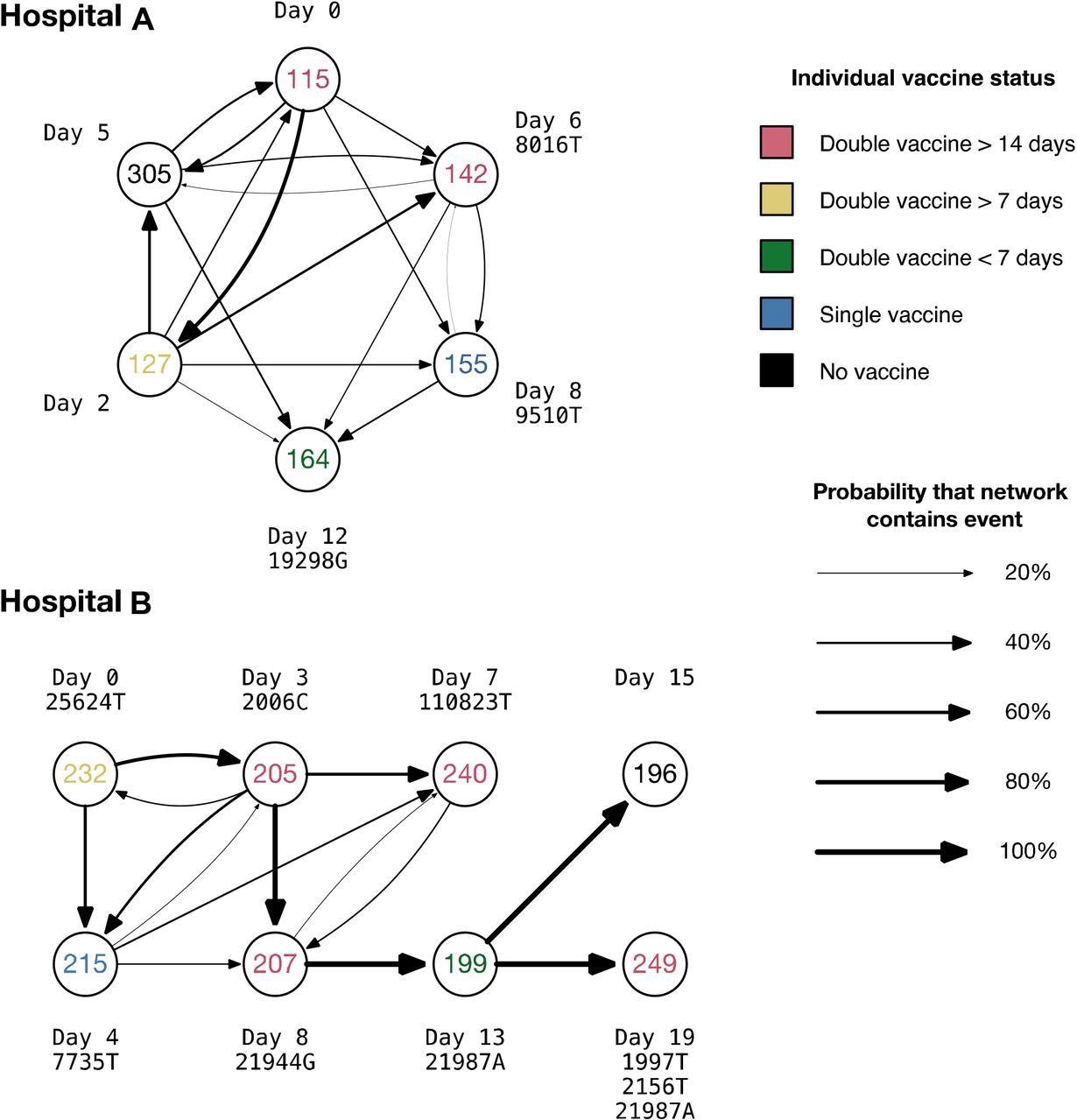
Notably, individuals 115, 127, and 305 in hospital A all had identical viral genomes. Overall, 1,381 possible transmission networks were recognized in hospital A, and 128 probable transmission networks were identified in hospital B, involving different medical staff.
When the data were compared to simulations, it was found that the transmission network inferred for hospital A is self-consistent. In contrast, the network inferred for hospital B is potentially distorted by missing data, with most individuals carrying unique mutations. A common genome sequence was shared by a junior medical staff member, a nursing student, and a nursing staff at hospital A – who developed illness over six days while three other team members had viral genomes that differed from this one a single SNP. In addition, nonsynonymous substitution T3082I in ORF1ab was observed in a junior medical staff member.
The present study has two significant caveats – firstly, it considered only symptomatic patients, excluding asymptomatic or people who failed to report infection status. Secondly, the reduced number of valid samples led to incomplete transmission chains.
Together, the present findings underscore the possibility of Delta variant transmission between fully vaccinated individuals in a healthcare setting, despite the high efficacy of vaccines in preventing severe disease. Furthermore, breakthrough infections were detected in hospital settings in people within 60 days of the second vaccine dose—when circulating neutralizing antibody levels were at their peak.
This study highlights the importance of continued infection control measures, even in highly vaccinated populations, and mask use in individuals who have received two doses of vaccination to reduce onward transmission.
*Important Notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Kemp, S., Cheng, M., Hamilton, W., et al. (2021), "Transmission of B.1.617.2 Delta Variant between vaccinated healthcare workers", medRxiv* preprint, doi: 10.1101/2021.11.19.21266406, https://www.medrxiv.org/content/10.1101/2021.11.19.21266406v1
Posted in: Medical Research News | Disease/Infection News
Tags: Adenovirus, Antibody, Bioinformatics, Coronavirus, Coronavirus Disease COVID-19, Cough, Efficacy, Fever, Genetic, Genome, Genomic, Healthcare, Hospital, Infection Control, Nucleotide, Nucleotides, Nursing, Phylogeny, Polymerase, Polymerase Chain Reaction, Respiratory, Reverse Transcriptase, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Vaccine, Virus

Written by
Nidhi Saha
I am a medical content writer and editor. My interests lie in public health awareness and medical communication. I have worked as a clinical dentist and as a consultant research writer in an Indian medical publishing house. It is my constant endeavor is to update knowledge on newer treatment modalities relating to various medical fields. I have also aided in proofreading and publication of manuscripts in accredited medical journals. I like to sketch, read and listen to music in my leisure time.
Source: Read Full Article
